Basic Raster Operations in R
Introduction
While much of urban data systems moved to vector object based data structures, raster datasets are ubiquitous and ever present. Raster datasets are simply an array of pixels/cells organized into rows and columns (or a grid) where each cell contains a value representing information, such as temperature, soil type, land use, water level. Rasters are aerial photographs, imagery from satellites, google street view images. Few things to note.
- Raster datasets are always rectangular (rows x col) similar to matrices. Irregular boundaries are created by using NAs.
- Rasters have to contain values of the same type (int, float, boolean) throughout the raster, just like matrices and unlike data.frames
- The size of the raster depends on the resolution and the extent of the raster. As such many rasters are large and often cannot be held in memory completely.
- In general, rasters we would interested in have a coordinate system (projected or unprojected). Because of it regular nature of the data structure, all we need to store is the Origin coordinates and the cell resolution (square or rectangular).
Rasters in R
The workhorse package for working with rasters in R is raster and terra packages by Robert Hijmans. In particular see Spatial Data Science with R. terra is better and faster in many instances, but is newer and does not have all the functionality and support associated with raster. I will switch back and forth between the two packages as necessary. Most functions are available in both packages, but it is useful to learn the differences
Eventually, if you get into the weeds in Remote Sensing, you should use RStoolbox package that has support for parallel processing. If you are working in rasters that are not geographically located (such as digital pictures) magick wrapper around ImageMagick may be a good option.
Download data
You can download the datasets used in this tutorial here
Reading in Rasters
Let’s start with toy examples and examine the use of functions in the raster library.
library(raster)
library(rasterVis)
library(sf)
library(tidyverse)
library(tigris)
library(leaflet)
library(skimr)
library(terra)
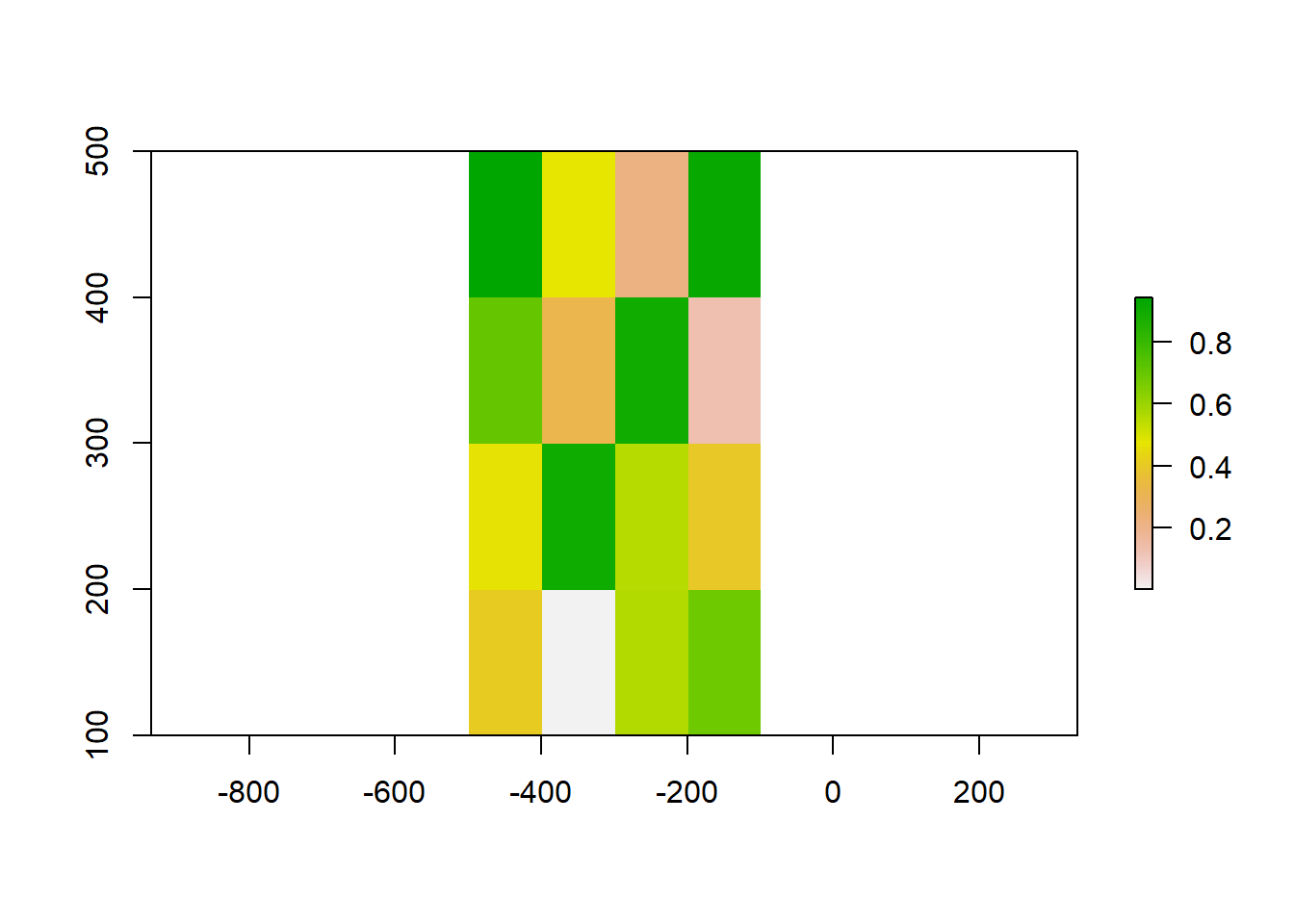
# Let's create a raster of size 5x5. #creates an empty raster object
raster_object <- raster(ncol=5, nrow=5, xmx=-100, xmn=-500, ymn=100, ymx=500, crs='+proj=longlat +datum=WGS84')
#In terra, you can create an object with 'rast'. Notice the difference between xmx and xmax argument names.
raster_object_terra <- rast(ncol=5, nrow=5, xmax=-100, xmin=-500, ymin=100, ymax=500, crs='+proj=longlat +datum=WGS84')
#view its properties
raster_object
# class : RasterLayer
# dimensions : 5, 5, 25 (nrow, ncol, ncell)
# resolution : 80, 80 (x, y)
# extent : -500, -100, 100, 500 (xmin, xmax, ymin, ymax)
# crs : +proj=longlat +datum=WGS84 +no_defs
raster_object_terra
# class : SpatRaster
# dimensions : 5, 5, 1 (nrow, ncol, nlyr)
# resolution : 80, 80 (x, y)
# extent : -500, -100, 100, 500 (xmin, xmax, ymin, ymax)
# coord. ref. : +proj=longlat +datum=WGS84 +no_defs
#access the resolution of the object
res(raster_object)
# [1] 80 80
#we can also set the resolution of the object
res(raster_object) <- 100
res(raster_object)
# [1] 100 100
# We can assign values to the object by supplying the right number of values.
values(raster_object) <- runif(ncell(raster_object))
plot(raster_object)
# Basic airthmetic is treated as
raster_object2 <- raster_object * 2
Exercise
- Notice the difference in the format between objects created by
rastandraster. Read up on those differences.
Of course, the real advantage is to reading in rasters that we can work with in the urban policy arena. Rasters come in many formats.
Say for example, we want to read in tree cover data in the ERDAS Imagine (*.img) format.
library(here)
(durham_treecover_raster <- here("tutorials_datasets", "basicraster", "durham_treecover.img") %>% raster)
# class : RasterLayer
# dimensions : 1483, 888, 1316904 (nrow, ncol, ncell)
# resolution : 30, 30 (x, y)
# extent : 1510245, 1536885, 1558875, 1603365 (xmin, xmax, ymin, ymax)
# crs : +proj=aea +lat_0=23 +lon_0=-96 +lat_1=29.5 +lat_2=45.5 +x_0=0 +y_0=0 +ellps=GRS80 +units=m +no_defs
# source : durham_treecover.img
# names : Layer_1
# values : 0, 100 (min, max)
# Download the boundary of the county for visualisation purposes.
durhamcty_sf <- counties(state = 37, refresh=TRUE, cb=TRUE, progress_bar = FALSE) %>%
st_as_sf() %>%
filter(GEOID == "37063") %>%
st_transform(crs = crs(durham_treecover_raster))
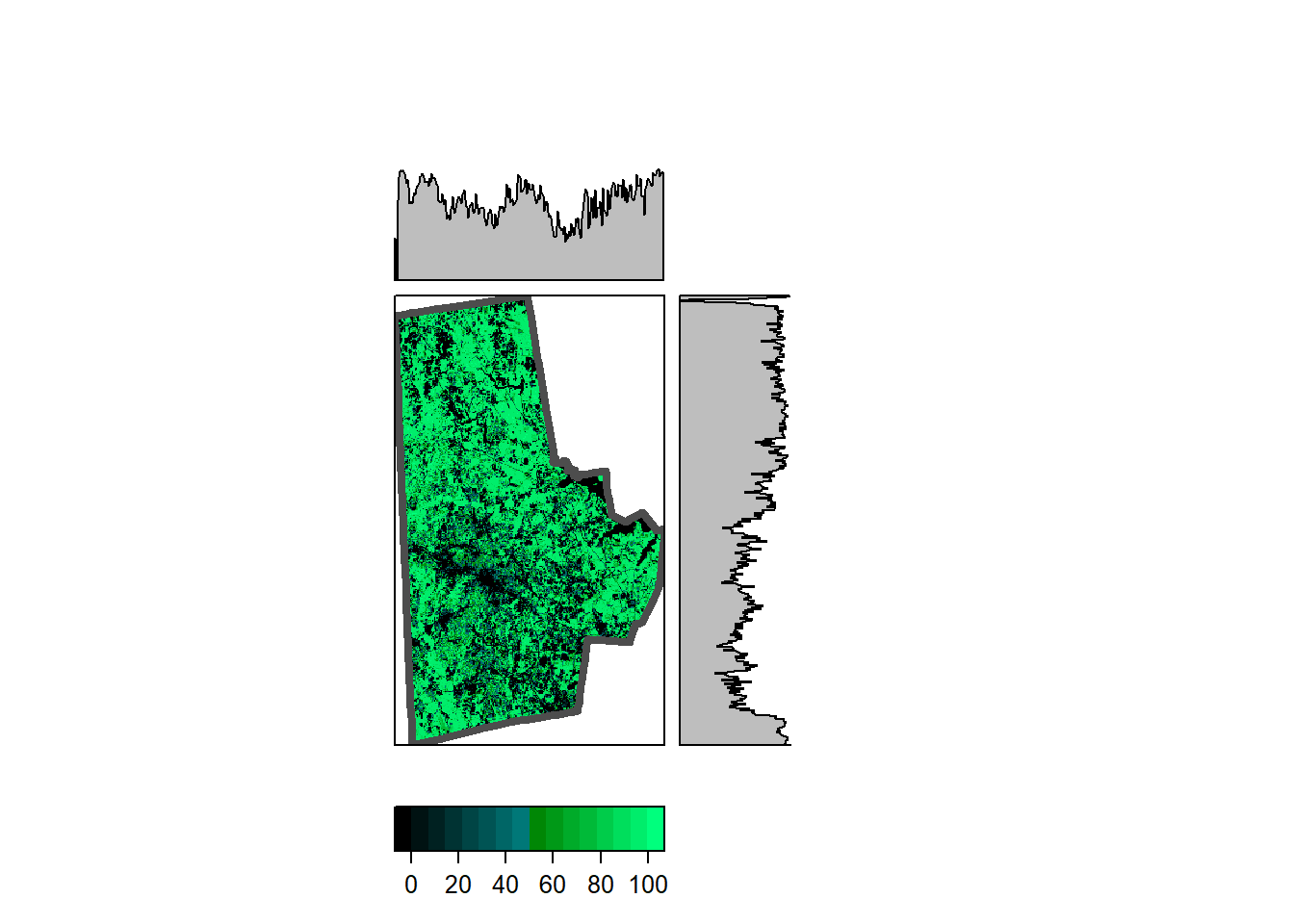
Note that the durham_treecover_raster only is a reference to a file and not loaded fully into memory. Also note the resolution, origin and dimensions. The tree cover is also an integer raster with values ranging from 0 to 100. Integers take up fewer bytes than floats and doubles, in general. The raster can be visualised as a static plot or in leaflet.
rasterVis::levelplot(durham_treecover_raster,
ylab=NULL, xlab=NULL,
scales=list(y=list(draw=FALSE), x=list(draw=FALSE)),
col.regions = rgb(0,0:100/100,0:50/100),
margin = list(FUN = 'median')
) +
latticeExtra::layer(sp.lines(as(durhamcty_sf, "Spatial"), col="gray30", lwd=4))
library(widgetframe)
leaflet() %>%
addRasterImage(durham_treecover_raster, layerId = "values", colors = colorNumeric("Greens", domain = 0:100)) %>%
leafem::addMouseCoordinates() %>%
# leafem::addImageQuery(durham_treecover_raster, type="click", layerId = "values") %>% # looks like broken in leafem.
frameWidget()
RasterLayer not SpatRaster.
In addition to visualisation we can process the entire raster or use a focal window. The first bit provides the median of all the values of the raster (removing NAs automatically) and the second creates a raster, whose value is the the median in 3x3 window around the focal cell. Note that we have treat NAs explicitly in the focal operations.
cellStats(durham_treecover_raster, "sd", asSample = FALSE)
# [1] 39.14223
durham_treecover_focal <- focal(durham_treecover_raster, w = matrix(1, nrow = 3, ncol = 3), fun = function(x,...){sd(x,na.rm=TRUE)})
# leaflet() %>%
# addRasterImage(durham_treecover_focal, layerId = "values", colors = colorNumeric("Greens", domain = 0:70)) %>%
# leafem::addMouseCoordinates() %>%
# # leafem::addImageQuery(durham_treecover_focal, type="mousemove", layerId = "values") %>% # Looks like this is broken in leafem. see https://github.com/r-spatial/leafem/issues/7
# frameWidget()
rasterVis::levelplot(durham_treecover_focal)
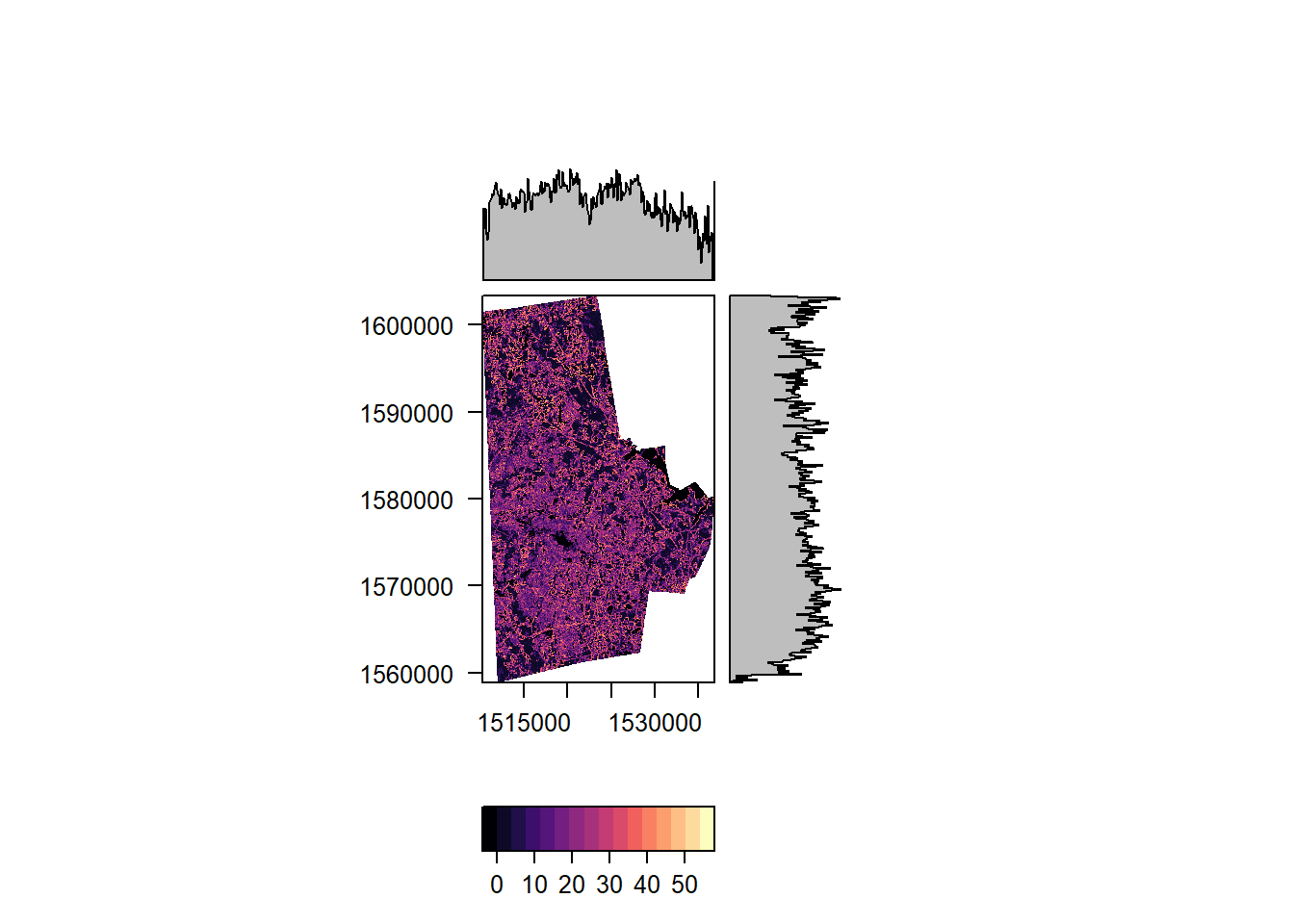
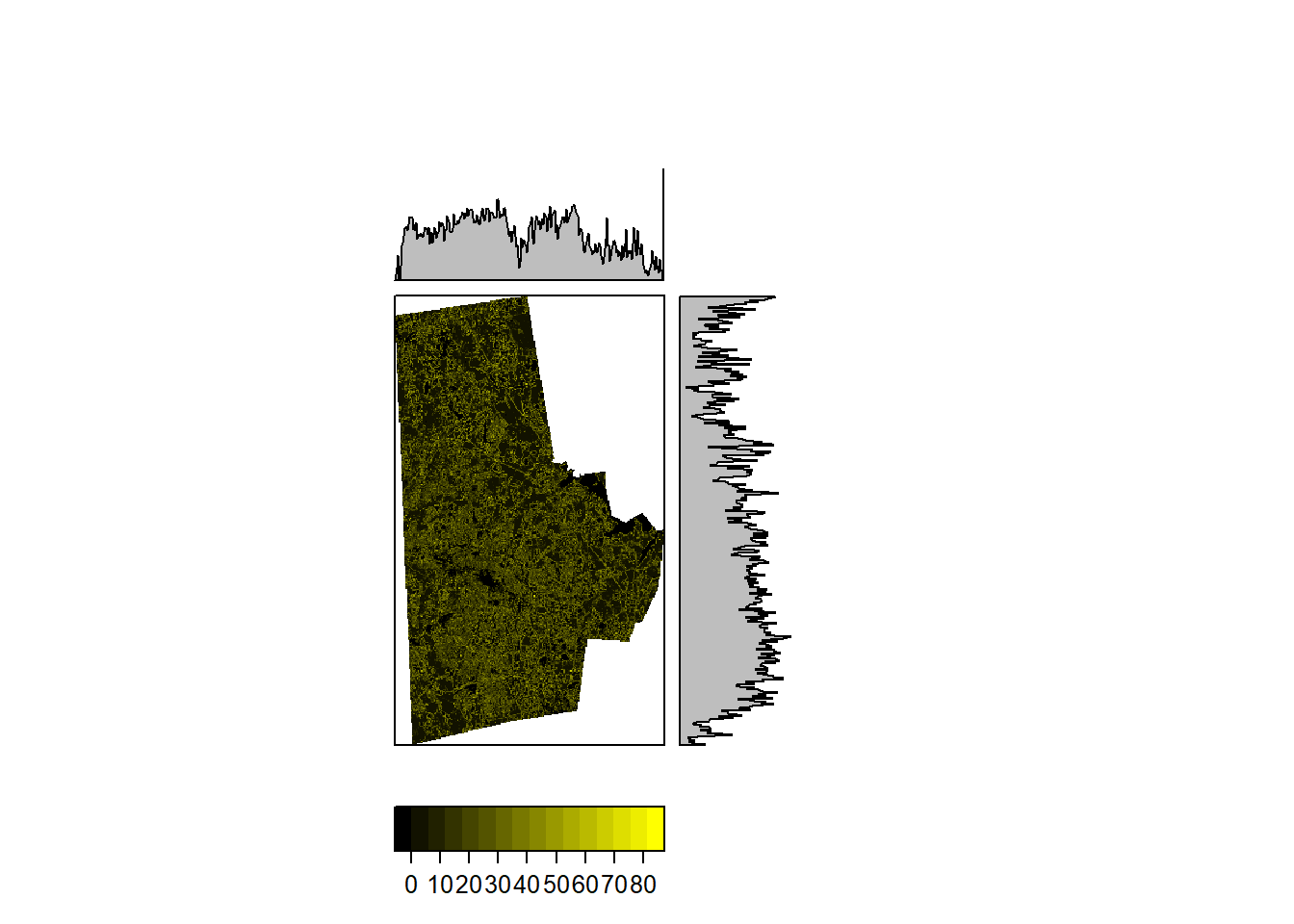
More complicated functions are certainly possible. The following takes the average of the deviation all the cells in the 3x3 neighborhood w.r.t. the value of the focal cell.
unevenness_raster <- focal(durham_treecover_raster, w=matrix(1, nrow=3, ncol=3), fun=function(x, ...) sum(abs(x[-5]-x[5]))/8, pad=TRUE, padValue=NA)
rasterVis::levelplot(unevenness_raster,
ylab=NULL, xlab=NULL,
scales=list(y=list(draw=FALSE), x=list(draw=FALSE)),
col.regions = rgb(0:100/100,0:100/100,0),
margin = list(FUN = 'median')
)
** Exercise **
- Use a different window (neighborhood) and calculate the focal values.
- Use a different function to summarize the values. How are cells on the edge treated, with lot of the neighbors being NAs?
Vectors & Rasters
In addition to raster based windows, we can use vector based neighborhoods and calculate the statistics (or any other metric). The only difference is that in a focal operation, the windows move over the raster cell by cell, where as zonal operations are exclusive, i.e. if a cell belongs to a zone, it is does not belong to another zone. For example, the following code calculates the Median and Interquartile ranges in each tract of Durham and stores them per tract.
This is where terra shines. I am going to convert the Rasterlayer to SpatRaster and proceed. However, terra requires the vector layer to be in its proprietary format. Fortunately, vect easily converts an sf object to a vect object.
durham_tracts_sf <- tracts(state='37', county='063', progress_bar = FALSE, year='2010') %>%
st_as_sf() %>%
st_transform(crs = crs(durham_treecover_raster))
durham_treecover_terra <- rast(durham_treecover_raster)
tree_bg <- terra::extract(durham_treecover_terra, vect(durham_tracts_sf))
tree_bg_summary <-tree_bg %>%
group_by(ID) %>%
dplyr::summarise(treecover_iqr = IQR(Layer_1, na.rm=T),
treecover_median = median(Layer_1, na.rm=T))
durham_tracts_sf <- cbind(durham_tracts_sf, tree_bg_summary) # Can do this because extract uses the record ID and we have not scrambled the sf object. Use with caution. See further down for a better method
durham_tracts_sf %>%
st_set_geometry(NULL) %>%
dplyr::select(treecover_median, treecover_iqr) %>%
skim
| Name | Piped data |
| Number of rows | 60 |
| Number of columns | 2 |
| _______________________ | |
| Column type frequency: | |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Table 1: Data summary
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| treecover_median | 0 | 1 | 46.20 | 24.92 | 0 | 31 | 45.5 | 65.5 | 92 | ▃▅▇▃▃ |
| treecover_iqr | 0 | 1 | 61.33 | 20.29 | 0 | 48 | 62.0 | 79.0 | 94 | ▁▂▇▇▇ |
Exercise
- Notice the output differences when you use
raster::extractandterra::extract.- Modify the code so that you can merge with the sf object.
- Run microbenchmarks on these two functions and see which is faster and more efficient. Also think about the collection of functions to get to the output and run the microbenchmarks. Bottlenecks can be in different places.
Categorical Rasters
Some times raster can be categorical (nominal or ordinal values), however, they are stored as numbers for convenience just as factors in regular R. For example a land cover raster might have a value 21 representing Developed Open Space and 95 representing Emergent Wetlands. While it is certainly possible to do arithmetic on them (additions and subtractions etc.), those operations do not make substantive sense. Explicit care should be taken to analyse these categorical rasters.
In particular, use ratify function to create a Raster Attribute Table and use it for analyses and visualisation as below.
durham_lc_raster <- here("tutorials_datasets", "basicraster", "c11_37063.img") %>%
raster() %>%
ratify()
nlcd_class <- here("tutorials_datasets", "basicraster", "legend_csv.csv") %>%
read_csv()
rat <- levels(durham_lc_raster)[[1]] %>% merge(nlcd_class, by.x="ID", by.y="Value")
levels(durham_lc_raster) <- rat
levelplot(durham_lc_raster, att="Classification", ylab=NULL, xlab=NULL, scales=list(y=list(draw=FALSE), x=list(draw=FALSE)), col.regions=levels(durham_lc_raster)[[1]]$colortable, main="2011 Land Cover")
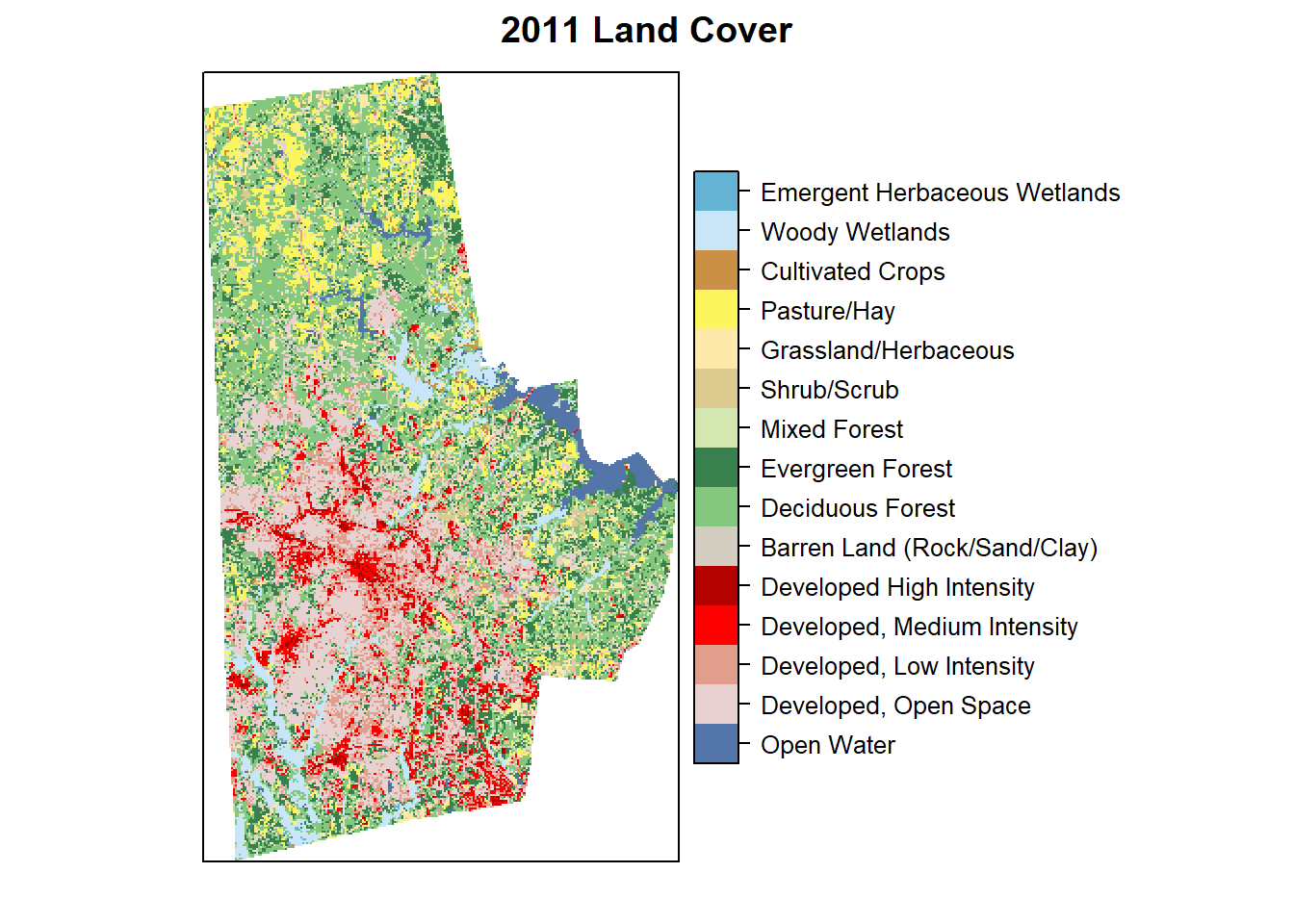
We can also extract all cells with in each polygon and produce counts using the table function. Confirm to make sure that the projections and vectors and rasters are the same.
proj4string(durham_lc_raster)
# [1] "+proj=aea +lat_0=23 +lon_0=-96 +lat_1=29.5 +lat_2=45.5 +x_0=0 +y_0=0 +ellps=GRS80 +units=m +no_defs"
crs(durham_tracts_sf)
# [1] "PROJCRS[\"Albers\",\n BASEGEOGCRS[\"GCS_GRS 1980(IUGG, 1980)\",\n DATUM[\"D_unknown\",\n ELLIPSOID[\"GRS80\",6378137,298.257222101,\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"Degree\",0.0174532925199433]]],\n CONVERSION[\"unnamed\",\n METHOD[\"Albers Equal Area\",\n ID[\"EPSG\",9822]],\n PARAMETER[\"Latitude of false origin\",23,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8821]],\n PARAMETER[\"Longitude of false origin\",-96,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8822]],\n PARAMETER[\"Latitude of 1st standard parallel\",29.5,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8823]],\n PARAMETER[\"Latitude of 2nd standard parallel\",45.5,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8824]],\n PARAMETER[\"Easting at false origin\",0,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8826]],\n PARAMETER[\"Northing at false origin\",0,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8827]]],\n CS[Cartesian,2],\n AXIS[\"(E)\",east,\n ORDER[1],\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]],\n AXIS[\"(N)\",north,\n ORDER[2],\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]]]"
durham_lc_terra <- rast(durham_lc_raster)
extract from terra conflicts with the same function from raster, so it is useful to explicitly specify the package the function comes from.
Furthermore, extract relies on on the record number or row number. It is useful to create a new table with the row number and an ID column like GEOID10 which can then be used to merge back.
tract_geoid <- tibble(ID = 1:nrow(durham_tracts_sf), GEOID10 = durham_tracts_sf$GEOID10)
lc_tracts <- terra::extract(durham_lc_terra, vect(durham_tracts_sf)) %>%
mutate(Layer_1 = paste0("lc", Layer_1)) %>% # This is an easy way to convert to characters and therefore to factors. Layer_1 and ID are columns in the tibble that results from from terra::extract
group_by(ID, Layer_1) %>%
summarize(count = n())
lc_tracts_tbl <- lc_tracts %>%
ungroup() %>% # Groups are no longer necessary
pivot_wider(names_from = Layer_1, values_from = count) %>%
left_join(tract_geoid)
lc_tracts_tbl
# # A tibble: 60 × 17
# ID lc11 lc21 lc22 lc23 lc24 lc41 lc42 lc43 lc52 lc71 lc81 lc90
# <dbl> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
# 1 1 35 3994 2233 790 176 1373 1139 224 91 175 72 7
# 2 2 9 4099 633 276 22 371 308 79 1 18 NA 854
# 3 3 NA 429 618 249 253 NA NA NA NA NA NA NA
# 4 4 3907 12486 2371 723 209 85032 25850 8729 5115 11342 39446 7882
# 5 5 NA 690 950 859 157 51 17 NA NA NA NA NA
# 6 6 9776 4085 755 412 76 25189 15132 4238 3421 5364 4986 1674
# 7 7 NA 2517 566 458 156 41 NA 35 NA 7 NA NA
# 8 8 914 5732 1114 212 5 12414 2242 919 565 1498 2801 1371
# 9 9 153 6603 731 108 NA 11765 3395 1670 381 952 1489 75
# 10 10 130 5958 1996 1387 224 3112 2519 410 105 369 308 3490
# # … with 50 more rows, and 4 more variables: lc31 <int>, lc82 <int>,
# # lc95 <int>, GEOID10 <chr>
Tips & Tricks
Exploit RasterOptions
There are two parameters that have a lot of influence on the performance of the package: chunksize and maxmemory.
- chunksize: integer. Maximum number of cells to read/write in a single chunk while processing (chunk by chunk) disk based Raster* objects
- maxmemory: integer. Maximum number of cells to read into memory.
You can override the default values using rasterOptions(maxmemory = 1e+09). Other options that are useful to set within rasterOptions are overwrite and datatype
Parallelise your operations
While rasters are large, you can take advantage of the fact that not all operations have to be sequential and can be done in parallel.
- Use
clusterRto parallelize some operations such ascalc,overlayandpredict. - Use
foreachanddoParallelpackages whenever possible.
Manage your temp files
When multiple operations are run on rasters, because they need to written to disk often, they create multiple copies of the same raster. This quickly takes up space and stops the script from moving forward. Use removeTmpFiles(h=) to periodically empty tempfiles. It is always a good idea to set temporary directory using tempdir() at the beginning of the script and then unlink() to delete the directory at the end of the script.
Conclusions
Rasters are incredibly useful in urban domains but are often left to remote sensing experts. In the next few tutorials we will see how they can be used to characterise urban form and identify suitable sites. They should sit along side with vector data manipulation in the analysts’ toolbox.
